DataJoint Elements
Type: Software,
Keywords: Data sharing, Data Management, Automation, Software Integration, Relational Databases, Scientific Workflows, Electrophysiology, Calcium Imaging, Pose Estimation
Resource ID: SCR_021894
Extensible workflows for neuroscience research
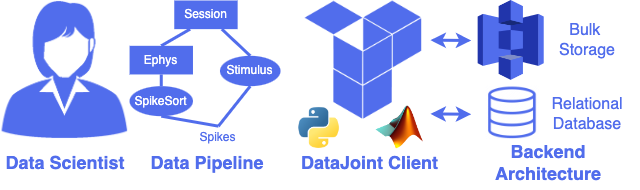
DataJoint Elements provides an efficient approach for neuroscience labs to create and manage scientific data workflows: the complex multi-step methods for data collection, preparation, processing, analysis, and modeling that scientists must perform in the course of an experimental study. The work is derived from the developments in leading neuroscience projects and uses the open-source DataJoint framework for interfacing databases and automating computations.
* Automates data ingestion and processing for neuroscience experiments.
* Supports modalities including extracellular electrophysiology, multiphoton calcium imaging, miniscope calcium imaging, pose estimation, and visual stimulation.
* Shared and extensible database programming for open science initiatives.
* Organizing data and computations for neurophysiology experiments end-to-end from data entry and acquisition to processing and visualization.
Each DataJoint Element is paired with an example workflow with Jupyter notebooks describing the setup and usage (e.g. https://github.com/datajoint/workflow-array-ephys).
* Flexible concurrent access to data.
* Data integrity
* Automated distributed computation.
* Collaborative analysis.
* Currently works only with MySQL-compatible database. Support for other database backends is planned in the future.
* Running DataJoint workflows requires configuring a database server and setting up file or object storage with access for all participants.
* Yatsenko, Dimitri, et al. “DataJoint Elements: Data Workflows for Neurophysiology.” bioRxiv (2021). doi: https://doi.org/10.1101/2021.03.30.437358.
Dimitri Yatsenko, CEO
DataJoint, Houston, TX
TEAM / COLLABORATOR(S)
Chris Brozdowski, Neuroscience Data Engineer, DataJoint
Dimitri Yatsenko, Principal Investigator, DataJoint
Kabilar Gunalan, Director of Neuroscience Research, DataJoint
Thinh Nguyen, Director of Neuroscience Projects, DataJoint
Raphael Guzman, Director of Engineering, DataJoint
FUNDING SOURCE(S)
NIH U24NS116470

