cytoNet
Type: Software,
Keywords: Image Analysis Software, Neural Networks, Cell Engineering, Cell-Cell Interaction, Graph Theory, Network Topology
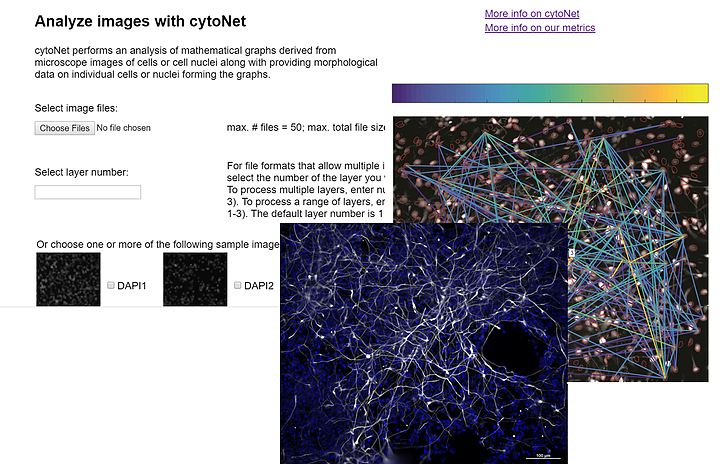
A mathematical, web-based tool to rapidly characterize multiscale networks from images
cytoNet provides a mathematical, web-based tool to rapidly characterize multiscale networks from images. To study complex tissue, cell and subcellular topologies, cytoNet integrates vision science with graph theory to quantify environmental effects on network topology. cytoNet applications include: (1) characterizing how pain sensation alters neural circuit activity in vivo, (2) quantifying patterns in how diverse brain cells respond to neurotrophic stimuli, & (3) uncovering cell cycle synchronization of differentiating neural stem cells. Awareness of cytoNet as a resource for the BRAIN Initiative community is a dissemination goal.
* Image analysis method
* Integrates visuals with graph theory to quantify environmental effects on network topology
* Identifies effects of cell-cell interactions on individual cell phenotypes
* Takes as input color or binary images of cells or tissues, and quantifies the spatial relationships in cell communities using principles of graph theory
* Great benefit in stem cell biology to evaluate environmental effects on cell fate decisions
* To rapidly characterize multiscale networks from images
* To quantify the structure of cell communities from microscope images
* To characterize how pain sensation alters neural circuit activity in vivo
* To quantify patterns in how diverse brain cells respond to neurotrophic stimuli
* To uncover cell cycle synchronization of differentiating neural stem cells
* To identify the spatial relationships between cells and to evaluate the effect of cell-cell interactions on individual cell phenotypes
* Used to study complex cell communities in a quantitative manner, leading to a deeper understanding of environmental effects on cellular behavior
* Used to quantitatively study biological pathways involved in cell-cell communication
* Characterize endothelial cell structures and differentiation of neural stem cells
* Characterize the morphological response of endothelial cells to neurotrophic factors present in the brain after injury
* Characterize cell cycle dynamics of differentiating neural progenitor cells
* Cloud-based image analysis software
* Web-based interface
* Easy to study complex tissue, cell and subcellular topologies
* The maximum number of files that can be uploaded at once onto cytoNet is 50, and the maximum total file size for a single upload is 100 MB. This limit can be circumvented by using the downloaded code.
Image files in JPG, PNG or TIF format
cytoNet: Spatiotemporal network analysis of cell communities. Mahadevan AS, Long BL, Hu CW, Ryan DT, Grandel NE, Britton GL, Bustos M, Gonzalez Porras MA, Stojkova K, Ligeralde A, Son H, Shannonhouse J, Robinson JT, Warmflash A, Brey EM, Kim YS, Qutub AA. PLoS Comput Biol. 2022 Jun 13;18(6):e1009846. doi: 10.1371/journal.pcbi.1009846. eCollection 2022 Jun. PMID: 35696439
Mahadevan et al. 2017, cytoNet: Network Analysis of Cell Communities, bioRxiv. doi: https://doi.org/10.1101/180273
https://www.qutublab.org/cytonet
https://www.biorxiv.org/content/10.1101/180273v1
Amina Ann Qutub, Cathy Burzik Professor in Engineering Design, University of Texas San Antonio
University of Texas, San Antonio
TEAM / COLLABORATOR(S)
Arun Mahadevan, Computational Neuroscientist, Rarebase
Byron Long, Assistant Professor, Computer Science, University of Texas San Antonio
Chenyue Wendy Hu, Data Scientist, Uber Technologies
Yu Shin Kim, UT Health Sciences
Kevin Francis, Sanford Research Institute
Eric Brey, University of Texas, San Antonio
Aryeh Warmflash, Rice University
FUNDING SOURCE(S)
* NSF CAREER 1150645
* NSF NCS 1533708
* NAKFI

