Cell Type Knowledge Explorer
Type: Software,
Keywords: Cell types, Taxonomy, Primary motor cortex, Transcriptomics, Patch-seq, Spatial transcriptomics, Single cell RNA-seq, Ontology, Epigenetics, Brain atlas
Resource ID: SCR_022793
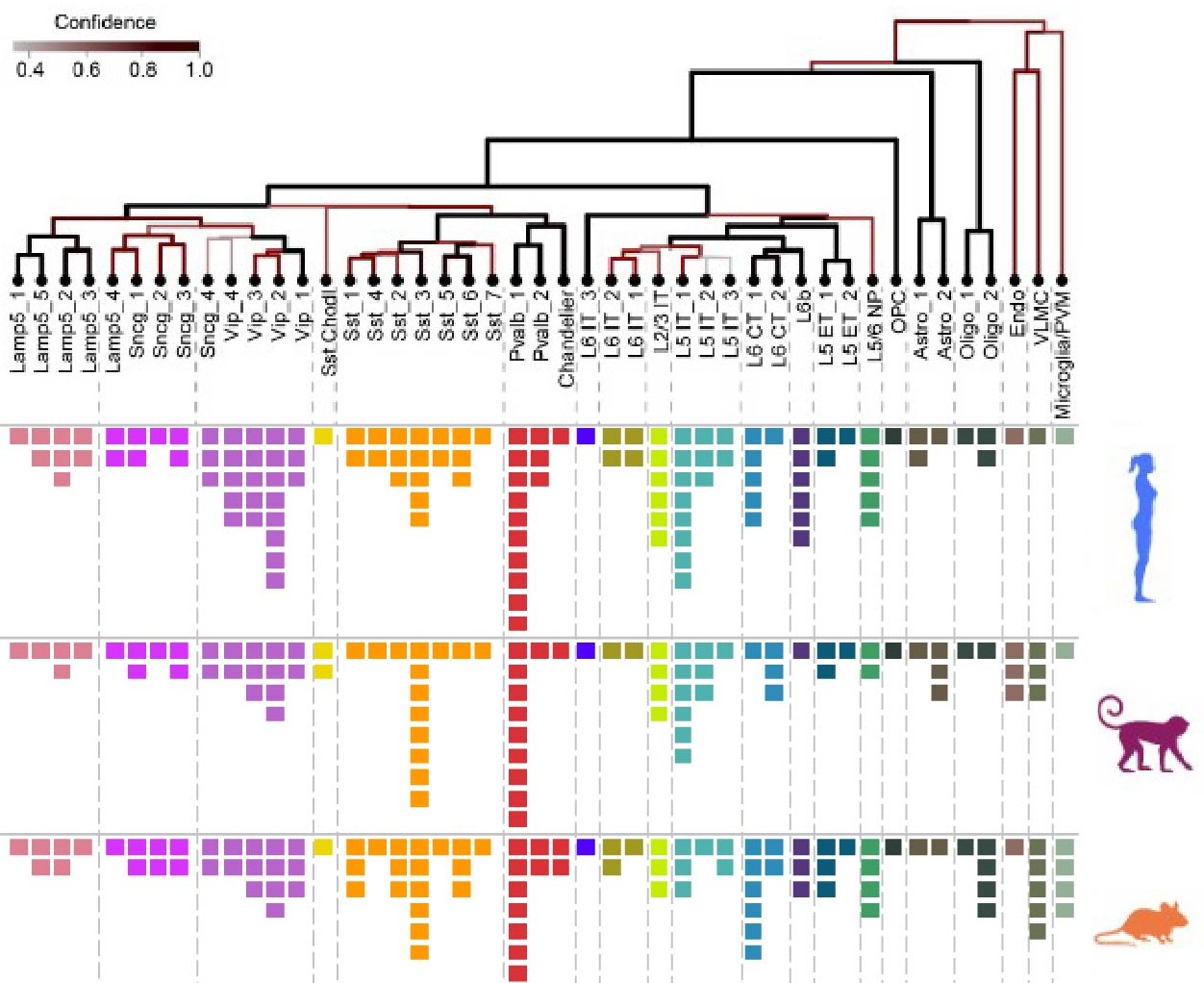
A web tool for exploration of cell types in primary motor cortex that spans data modalities, species, and levels of resolution
The Cell Type Knowledge Explorer (CTKE) is a publicly accessible web application to browse data associated with the multimodal cell census and atlas of primary motor cortex, developed in close collaboration with the BRAIN Initiative Cell Census Network (BICCN). The CTKE includes access to single-cell transcriptomic and epigenomic profiling of human, marmoset, and mouse primary motor cortex, with additional data sets assessing the spatial distribution, cell morphology (shape), and electrical properties of these cell types. All data powering the CTKE is provided to encourage cell exploration by scientists, students, and educators through the browser and linked resources.
* Unique tool for visualization of defining features of individual cell types in primary motor cortex across several modalities simultaneously.
* Provides easy access to underlying BICCN data sets and related data exploration tools such as epigenetics browsers and tools for cell type mapping.
* Comparison of matched cell types across mouse, human, and marmoset via a web portal.
* Ontology engine drives a cell type search and connects these data to external knowledge about cell types.
* The CTKE teaches researchers and students about the defining features of different cell type in primary motor cortex, and how these cell types compare with one another and with matching types across species.
* The CTKE helps researchers to annotate and interpret their own data, by comparing their own data computationally (e.g., mapping single cell RNA-seq data) or visually (e.g., comparison of spatial location or cell morphology) with presented cell types.
* The CTKE provides a jumping point to access many additional data sets and external tools for exploring cell types in primary motor cortex.
* The CTKE helps researchers to annotate and interpret their own data. For example, CTKE includes links to Azimuth, a web application that provides utilities to map single-cell expression data to curated reference datasets. This allows users to derive cell type annotations for their own datasets in the context of BICCN primary motor cortex mini atlas data for the human and mouse. Similarly, CTKE facilitates the interpretation and annotation of other data types. For example, a researcher studying mouse MOp may have immunohistochemistry data indicating that the gene Rorb is highly expressed in a certain population of cells and want more information about what type of cells they might be. Searching “Rorb” in the CTKE would return the L4/5 IT neuron subclass as a cell type that expresses Rorb more highly than other MOp types. Navigating to the “Spatial Transcriptomics” and “Morphology” panels would reveal that L4/5 IT neurons are found at a similar cortical depth and with similar morphological characteristics to those this researcher sees in their cell population of interest. If this researcher were interested in understanding whether these cell types are present in humans, they could navigate to the “Cross-Species Cell Types” panel on the Cell Type Knowledge Card for the L4/5 IT_1 subtype, where they would also find several putatively homologous types and be able to navigate directly to their cards for further investigation.
* The CTKE helps student researchers learn about brain cell types. As single-cell transcriptomics technology advancements have brought new ways to characterize cell types, this knowledge must be conveyed to the next generation in an approachable manner. The CTKE visualization shows the three hierarchical classifications of the cell types (class, subclass, and cell types) and how within cell types it is defined by transcriptomics but contains other modalities (electrophysiological and morphology) in an approachable manner. We have created a 200/300-level undergraduate level lesson explaining the concept of cell types and have students use the CTKE to explore individual cell types further. The lesson has been used at Middlebury College in the Fall of 2023 and planned to be used at Western Washington University in the Spring of 2024. The finalized public version of the lesson will be released by the end of 2024 on our education page (https://alleninstitute.org/education/).
* The CTKE allows researchers to compare their disease models to a standard control. For example, a researcher studying a mouse model of a disease that affects the MOp (i.e., ALS), may do electrophysiological recordings of cells to assess how cells are impacted by disease. After analysis, researchers might notice a difference in the electrophysiological properties (firing patterns, input resistance, firing rate, AP amplitude and/or width, etc.) of their disease model L5 ET cells compared to the healthy control cells listed in the CTKE. To gain a better understanding of which cell types within the L5 ET cell subclass are most affected, researchers can analyze the data provided within CTKE. For example, the L5 ET cell subclass contains four cell types, L5 ET_1, L5 ET_2, L5 ET_3, and L5 ET_4, which express different marker genes. CTKE data show that Npsr1 is a marker gene for L5 ET_4, but not for the other three cell types. Therefore, researchers could perform immunohistochemical staining with an NPSR1 antibody to determine if the cells affected in their disease model specifically belong to the L5 ET_4 cell type. Afterwards, additional immunohistochemical staining could be done for marker genes for the other three L5 ET cell types to determine if their disease model has cell type specific effects or if all L5 ET cells are affected indiscriminately.
* Human
* Marmoset
* Mouse
* This is a first-of-its-kind application that summarizes information about specific cell types across multiple modalities from several publications in a single place.
* This tool does not link to a cell database and therefore is not extensible to other brain regions and cell types in its current form; however, components of this tool with high importance or popularity may be integrated into the Allen Brain Cell (ABC) Atlas (RRID: SCR_024440) later.
* Exploration of the full feature space is possible only through connection to external tools.
* Internet connection
BRAIN Initiative Cell Census Network (BICCN), 2021, A Multimodal Cell Census and Atlas of the Mammalian Primary Motor Cortex, Nature, https://doi.org/10.1038/s41586-021-03950-0
Cell Type Knowledge Explorer tutorial, https://www.youtube.com/watch?v=xJRBQzZLX4s
GitHub repository with scripts and input files for creating web content, https://github.com/AllenInstitute/CTKE_viz
GitHub repository for back-end cell type ontology, https://github.com/obophenotype/brain_data_standards_ontologies
Allen Brain Map Community Forum, https://community.brain-map.org/
Media: August 2023 article on “The Quest to Discover the Cell Types”, https://www.technologynetworks.com/cell-science/articles/the-quest-to-discover-the-cell-types-of-the-brain-378272
Award: DataWorks! “Exemplary Achievement Award for Data Reuse”, https://datascience.nih.gov/director/directors-blog-dataworks-winners-2023
Past workshop: “Navigation and applying cell type taxonomies and tools from the Allen Institute for Brain Science” at IBRO 2023, https://alleninstitute.org/events/ibro2023/
Past workshop: “Satellite Event: Open resources for cell types and taxonomies with the Allen Brain Map” at SfN 2023, https://alleninstitute.org/events/sfn2023/
Future workshop: “Describe Your Neurons Like the Allen Institute” at the Allen Institute April 2024, https://alleninstitute.org/events/describe_your_neurons/
Future webinar series: “Cell Type Taxonomies A-Z”” in 2024 with CTKE the main topics in March and April 2024 (will be recorded for later viewing), https://alleninstitute.org/events/cell_type_az_webinars/
Mike Hawrylycz, Investigator
Allen Institute for Brain Science, Seattle WA<br />
TEAM / COLLABORATOR(S)
Bosiljka Tasic, Director of Molecular Genetics, Allen Institute for Brain Science
Brian Long, Scientist III, Allen Institute for Brain Science
Brian Staats, Associate Director of Data Visualization and Application, Allen Institute for Brain Science
Carol Thompson, Associate Director of Data Management, Allen Institute for Brain Science
Chris Mungal, Staff Scientist and Department Head of Biosystems Data Science, Lawrence Berkeley National Laboratory
Christine Lin, Scientific Data Engineer I, Allen Institute for Brain Science
Christof Koch, Meritorious Investigator, Allen Institute
Cindy van Velthoven, Associate Investigator, Allen Institute for Brain Science
David Osumi-Sutherland, Principal Research Scientist, Wellcome Trust Sanger Institute
Ed Lein, Senior Investigator, Allen Institute for Brain Science
Hongkui Zeng, Executive Vice President & Director of Allen Institute for Brain Science, Allen Institute for Brain Science
Hüseyin Kir, Research Scientist, Wellcome Trust Sanger Institute
Jeremy Miller, Senior Scientist, Allen Institute for Brain Science
Julie Nyhus, Principal Scientific Project Coordinator, Allen Institute for Brain Science
Kaitlyn Casimo, Manager of Education & Engagement, Allen Institute
Katherine Baker, Scientific Data Engineer I, Allen Institute for Brain Science
Katie Glattfelder, Research Administrator II, University of California Davis
Lane Sawyer, SW Engineer III, Allen Institute for Brain Science
Lauren Alfiler, Education Program Specialist III, Allen Institute
Luke Esposito, Sr. Director of Scientific Operations, Allen Institute for Brain Science
Lydia Ng, Investigator, Allen Institute for Brain Science
Maryann Martone, Professor Emerita, University of California San Diego
Mike Hawrylycz, Investigator, Allen Institute for Brain Science
Nathan Gouwens, Assistant Investigator, Allen Institute for Brain Science
Nomi Harris, Program Manager, Lawrence Berkeley National Laboratory
Pamela Baker, Scientific Data Engineer II, Allen Institute for Brain Science
Patrick Ray, Ontologist, Allen Institute for Brain Science
Prajal Bishwakarma, Scientific Data Engineer III, Allen Institute for Brain Science
Rachel Dalley, Morphology Manager, Allen Institute for Brain Science
Rachel Hostetler, Scientist I, Allen Institute for Brain Science
Raymond Sanchez, Product Manager II, Allen Institute for Brain Science
Richard Scheuermann, Scientific Director at the National Library of Medicine, National Institutes of Health
Rohan Gala, Scientist II, Allen Institute for Brain Science
Saroja Somasundaram, Bioinformatics II, Allen Institute for Brain Science
Shawn Tan, Senior Research Data Steward, Novo Nordisk
Shoaib Mufti, Senior Director of Data and Technology, Allen Institute for Brain Science
Susan Sunkin, Director of Scientific Program Management, Allen Institute for Brain Science
Tim Fliss, Software Engineer III, Allen Institute for Brain Science
Tina Ruiz, Executive Assistant, Allen Institute for Brain Science
Tom Gillespie, Knowledge Engineer, University of California San Diego
Trygve Bakken, Assistant Investigator, Allen Institute for Brain Science
Tyler Mollenkopf, Associate Director of Product Management, Allen Institute for Brain Science
Yasmeen Hussain, Scientific Project and Alliance Manager, Allen Institute for Brain Science
Yun (Renee) Zhang, Assistant Professor, J Craig Venter Institute
Zizhen Yao, Assistant Investigator, Allen Institute for Brain Science
FUNDING SOURCE(S)
* NIH RF1MH123220-01
* NIH U24NS133077-01

