Many people believe that the future lies in open science. Open science broadly refers to efforts to make scientific research accessible to all levels of society, from amateurs to professionals. Thanks to the creation of openly available brain databases by Brain Research through Advancing Innovative Neurotechnologies (BRAIN) Initiative® investigators — this future is quickly becoming a reality. Projects focused on accelerating machine learning, large-scale data storage, and cooperative brain and cell-type mapping are helping to set a new precedent for the integration of open science into thriving research programs across the country.
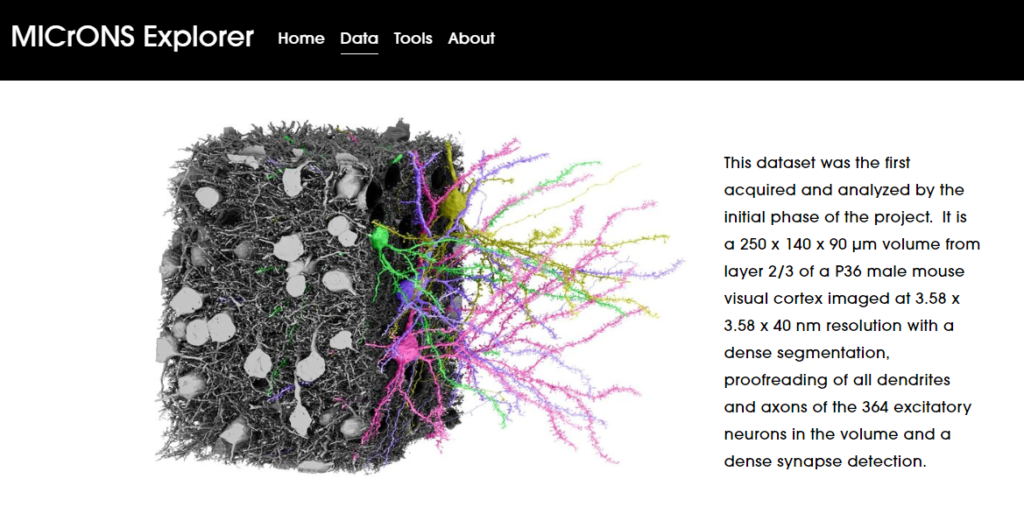
The open-access dataset that can be explored through the MICrONS program is focused on the cortical neurons (layer 2/3) from the mouse visual cortex.
Machine Intelligence from Cortical Networks (MICrONS)
The program known as Machine Intelligence from Cortical Networks (MICrONS) is advancing the field of machine learning by reverse-engineering the algorithms of the brain. Launched by the Intelligence Advanced Research Projects Activity (IARPA) in the Office of the Director of National Intelligence, MICrONS combines neuroscience and data science to advance machine learning by uncovering how the cortex performs computations at the mesoscale. The key players in this program are found at the Allen Institute, Baylor College of Medicine, and Princeton University. Together, IARPA investigators and their collaborators recently launched a public observatory of connectomics data that they generated through the MICrONS program. This was accomplished using large-scale electron microscopy. These data include reconstructions of neural circuits from the mouse visual cortex, as well as corresponding functional imaging data from the same cortical neurons. On the MICrONS website, even an amateur can explore electron microscopy data from layer 2/3 of the mouse visual cortex using an online visualization tool called Neuroglancer.
The MICrONS program has also made a splash in the world of academia. The most recent of the scientific papers associated with MICrONS are readily available on preprint servers such as bioRxiv, including this recent paper from the Seung lab at Princeton University on the variation of synapses between cortical pyramidal neurons. In fact, the Seung lab is quite excited about MICrONS and the future of open science. Lab member and Ph.D. candidate, J. Alexander Bae, had this to say:
Our lab is an untiring advocate of open science. Open science enables many perspectives to converge on the major scientific questions of our time. It ensures the highest-quality research, and inspires newcomers to the field. We think this is especially helpful for our work as the scope of the challenge is massive – extracting all of the scientific value from these datasets is simply too great a challenge for any one research team…The MICrONS program was a special opportunity to contribute to labs that otherwise might never access such data. As a student with an international background, I know that not all labs have the resources afforded by the MICrONS program, especially in countries outside of the extremely well-developed research environments in the United States. By releasing our data to the public (MICrONS Explorer), this gap can be minimized, and everyone in the community can access this data to the benefit of the entire field. I believe open science can accelerate the development of neuroscience as it creates an environment where this borderless communication between researchers is possible. – J. Alexander Bae, Princeton Neuroscience Institute
The Seung team is one of several contributing to the future of open science with the help of MICrONS; now is an exciting time to be involved with such a program.
Block and Object Storage Service (bossDB)
In the same vein, the Block and Object Storage Service (bossDB) uses an information system developed through MICrONS and is now deployed in support of the BRAIN Initiative to openly host a wide variety of large-scale volumetric neuroscience imaging datasets. The bossDB database lives in the Amazon Web Services cloud server. Here, investigators can scalably and securely store petabytes (millions of gigabytes) of high-dimensional data. Like MICrONS, these data are readily available to be explored by the research community. For instance, Dr. Josh Morgan and his team uploaded a dataset to bossDB this year that includes electron microscope images of the mouse dorsal lateral geniculate nucleus. Now that this data has been uploaded to bossDB, there are several ways that anyone can interact with it. To illustrate, people can launch the dataset with Neuroglancer, similar to how one can interact with the MICrONS datasets. However, bossDB provides even more options for data processing and visualization via a suite of tools that are available on their website. Each project may have different options. Some, for example, provide the raw data and annotations in the form of archived files that can be analyzed in Python, while other projects have links to Mendeley Datasets where Microsoft Excel files can be found. In short, bossDB provides a new cloud-native service for extremely large datasets. It also provides support for data ingest, storage, visualization, and sharing through a RESTful Application Programming Interface (API) and web-based user interface.
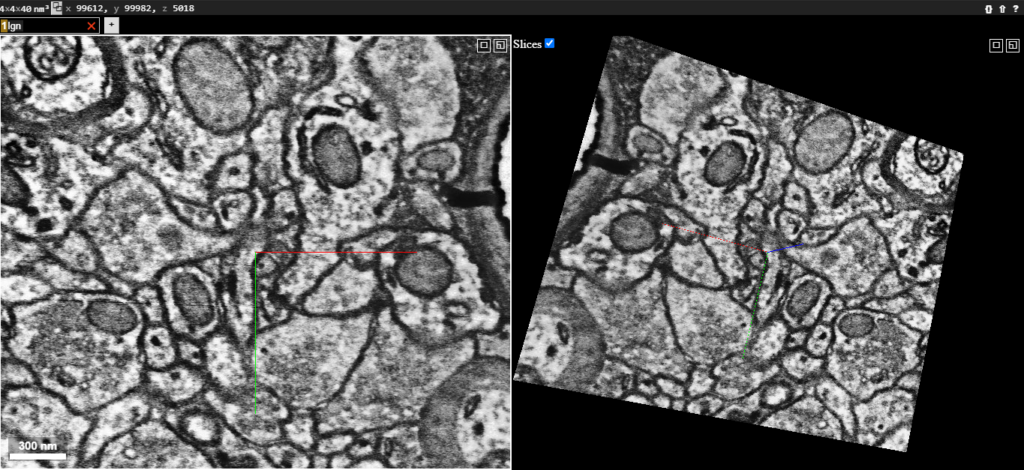
Visualizing the Morgan et al. 2020 electron microscopy data on bossDB using Neuroglancer.
Allen Institute for Brain Science
The Allen Institute for Brain Science, another active member of the BRAIN Initiative, also promotes open science. One of their open science projects is called the Allen Brain Map. This project has now made available many different datasets across species. For example, the Allen Cell Types Database is a comprehensive collection of cortical cells from both humans and mice, including standardized morphology, electrophysiology, and transcriptomics data. Another project of the Allen Institute is the Allen Mouse Brain Connectivity Atlas, which provides a mesoscale map of the whole mouse brain and integrated volumetric maps of neural pathways for all of the major anatomical areas and cell classes within the mouse brain. One more exciting component of this institute is the Allen Brain Observatory, which is a repository of visual coding datasets. This comprehensive database of cortical cell activity was collected through in vivo recordings using both 2-photon imaging and high density Neuropixels probes. Dr. Hongkui Zeng, Executive Vice President and Director of the Allen Institute for Brain Science says:
What I am most excited about is our ability to set a long-term vision, ask what are the most critical questions we could address and how we can serve the community in the best way, and then develop a series of projects around these goals and go for them as a team together. All these aspects are a wholesome package, it elevates the meaning of our everyday work. For example, we have been working on the problem of cell types – the parts list of the brain – using a comprehensive set of experimental approaches. “What is a cell type?”, is a fundamental question in biology. Understanding it and uncovering all the cell types in the brain lays the foundation for understanding how the brain works in health and disease. We are generating amazing datasets for the community and also learning underlying principles of brain organization. With these kinds of larger goals in mind, I find our work incredibly rewarding and fulfilling. – Hongkui Zeng, Ph.D., Executive Vice President and Director of the Allen Institute for Brain Scienc
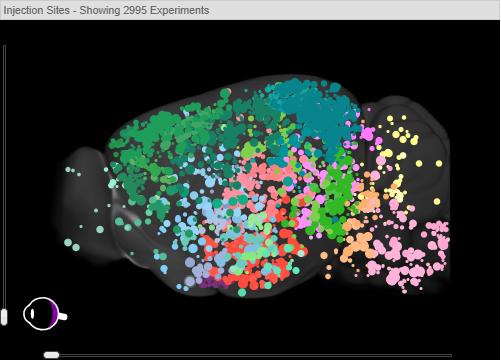
An image from the Allen Mouse Brain Connectivity Atlas that shows the injection sites from nearly 3000 experiments.
BRAIN Initiative Cell Census Network (BICCN)
The BRAIN Initiative Cell Census Network (BICCN), funded by the National Institutes of Health (NIH)’s BRAIN Initiative, is another program working to create a future that is characterized by open science and data sharing by creating an open-access digital brain atlas system. The overarching goal of the BICCN is to generate comprehensive three-dimensional common reference brain cell atlases. Another of the BICCN’s goals is to use these atlases to integrate molecular, anatomical, and functional data in order to classify and describe different brain cell types in several model systems, including mice, non-human primates, and humans. The BICCN is also a cooperative network that promotes collaboration among projects within the BRAIN Initiative that are geared toward creating these comprehensive brain atlases. One BICCN funded research project working toward this goal recently published a paper that focused on using 7 Tesla magnetic resonance imaging (MRI) on human brain tissue ex vivo to visualize the three-dimensional neuroanatomy of human brain in ultra-high resolution. According to one of the senior authors of this study, Dr. Bruce Fischl:
A central goal of our BICCN grant is to develop acquisition and analysis technology to facilitate the construction of a surface-based laminar and architectonic coordinate system in which to instantiate cell-census results. The 100µm dataset is an example of this type of technology, which we believe will facilitate a greater understanding of the mesoscopic structure of the human brain as well as improve the interpretation of numerous in vivo imaging studies. – Bruce Fischl, Ph.D., Professor, Harvard University
BRAIN-funded investigators such as these BICCN researchers will help form a comprehensive atlas of the human brain, which will greatly move the open science mission forward.
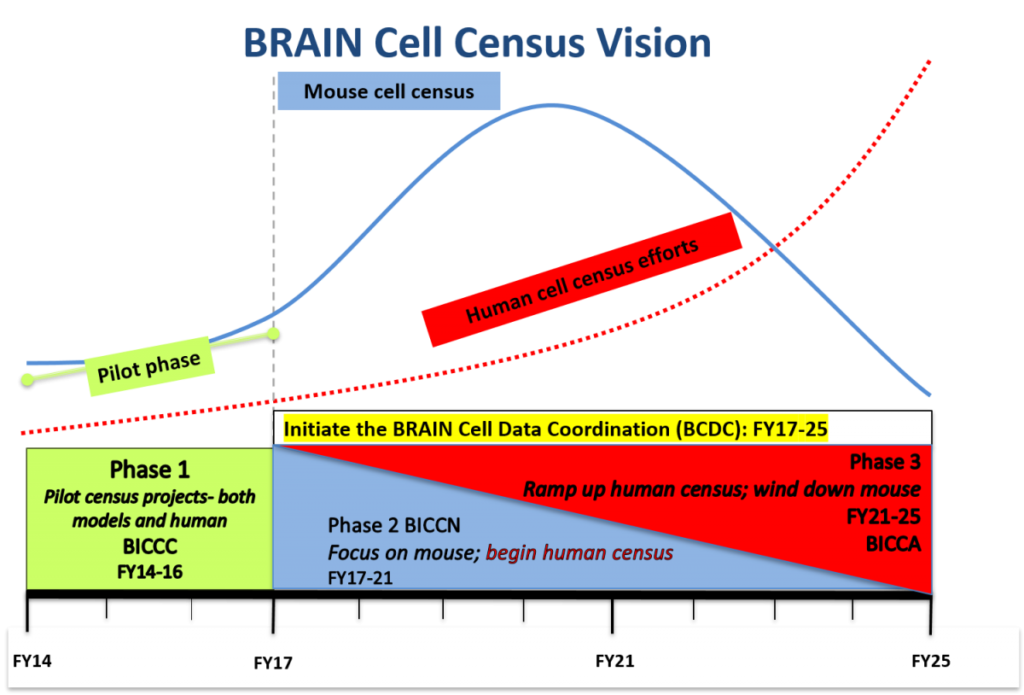
The BICCN Vision depicted as a timeline of events during the funding period.
Looking Ahead
The open availability of large databases represents a major milestone in the field of neuroscience. In fact, one could even say that these large, transparent, and accessible open science databases are the future of neuroscience, as well as other fields of science. The new director of the NIH BRAIN Initiative, Dr. John Ngai, had this to say about these programs and the future of data accessibility:
Research funded by the NIH BRAIN Initiative has been developing experimental tools and data resources at an impressive pace, one that I expect will accelerate with recent advances in technologies in biological imaging, physiology, and genomics. Dissemination and democratization of neurotechnologies and data resources represent core values of the BRAIN Initiative. We have established a robust and growing data infrastructure to ensure that data resources generated by BRAIN Initiative-supported projects will be accessible to the community at large for the benefit of the scientific enterprise.- John Ngai, Ph.D., Director, NIH BRAIN Initiative
Without a doubt, we are living in intriguing times. Moreover, we should be excited for the things to come and the future of open science, particularly those that will result from the efforts of the BRAIN Initiative and its current and future investigators.

